Abstract
Introduction: All-trans retinoic acid (ATRA, RA) has powerful activity in acute promyelocytic leukemia (APL); its efficacy in non-APL acute myeloid leukemia (AML) is still unclear, but may be enhanced by epigenetic drugs such as azanucleoside DNMT inhibitors (Blagitko-Dorfs et al. PLoS ONE 2013). In a randomized phase II study (DECIDER trial, NCT00867672) the addition of RA to decitabine (DAC) in newly diagnosed non-fit older AML patients resulted in a clinically meaningful extension of survival. We hypothesize that the add-on of RA to DAC results in cooperative transcriptome changes (possibly associated with demethylation) which may, at least in part, explain these clinical results.
Materials and Methods: U937 cells were treated in triplicates with daily pulses of 200 nM DAC, 1 µM RA was administered after 48 hours (hr). Cells were harvested after 72 and 120 hr of treatment. Proliferation and apoptosis were determined by acridine orange / propidium iodide staining. For the colony formation assay (CFA), 100 cells were seeded after 120 hr treatment in methylcellulose, colonies were counted after 21 days. Stranded cDNA libraries were sequenced on an Illumina HiSeq2000 with 60 million 125 bp paired-end reads. Read quality was checked with FastQC (per base Q-score >38), alignment to the GencodeCompV24 reference genome with RNAStar, generation of count matrices with ht-seq and differential expression testing with DeSeq2. Transcript changes were considered significant with an adjusted FDR p-value < 0.01. GO analysis was performed with Metascape. Methylomes were generated using Infinium Human Methylation 450K BeadChip arrays and differential methylation data was obtained using the RnBeads package for R (Assenov et al. Nature Methods 2014).
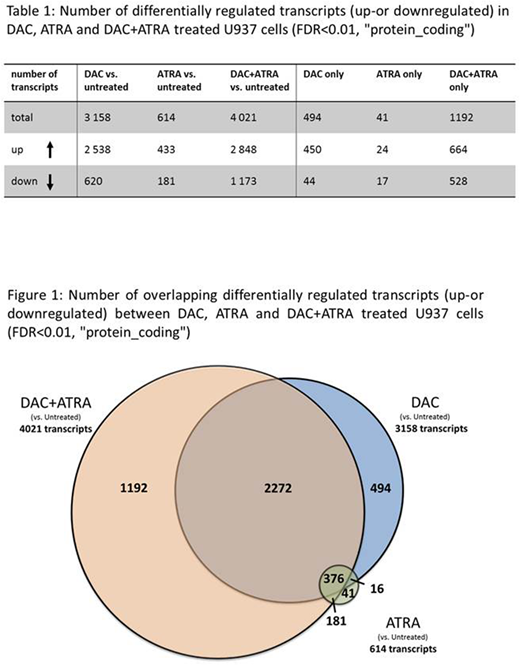
Results: In suspension culture, RA alone had no effect on the proliferation of U937 cells, DAC alone reduced proliferation by 74 %, the DAC+RA combination by 81 % (with viability always ≥ 90 %). RA alone also had no effect on colony growth, DAC reduced colony formation by 85 %, and DAC+RA decreased colony numbers by 96 %. To determine transcriptional add-on effects of DAC+RA, transcriptomes of cells treated for 72 hr were analyzed by RNA-Seq. RA alone had a modest effect on transcription, whereas DAC alone induced / downregulated 2538 and 620 transcripts, DAC+RA induced / downregulated 2848 and 1173 transcripts, respectively (Table 1, Fig. 1). 29.6 % (1192 transcripts) of all DAC+RA-altered transcripts were uniquely regulated by the combination and not by DAC or RA alone. GO analysis of the DAC+ATRA alone upregulated genes (664 transcripts) revealed a significant enrichment for immune system and cellular decay terms, downregulated genes were enriched for transcription and cell cycle terms. Of the transcripts already up- or downregulated by DAC or RA alone, 55.5 % of were further enhanced by DAC+RA. Among those, the retinoic acid receptors RARA, RARB and the retinoic acid response element (RARE)-containing tumor suppressor HIC-1 (Hassan et al. Cell Reports 2017) were induced 8.6-, 8.3- and 179.8-fold, respectively, compared to 2.4- and 5.3-fold (RARA and RARB, DAC alone) and 26.5-fold (HIC-1, RA alone) (validated by qRT-PCR). Among the most regulated uniquely induced transcripts by DAC+RA, the RA-regulating cytochrome P450 member CYP26A1 (regulated by multiple RAREs) was upregulated 347.3-fold.
In order to further investigate this add-on effect of RA on DAC-induced transcriptome changes, DNA methylation was analyzed (72 and 120 hr). As expected, DAC significantly reduced CpG methylation in gene bodies and promoters, without further demethylation upon RA add-on after 72 hr (after 120 hr, a trend towards further demethylation was noted with DAC+ATRA).
Conclusion:In vitro, the combination of DAC with RA resulted in enhanced growth inhibition and reduced colony formation compared to DAC alone. Transcriptome analysis revealed a high number of uniquely regulated transcripts, and an enhancement of regulation by the combination treatment. As expected, this also included genes with RAREs. The enrichment for genes involved in immune response (e.g. multiple interferon-response genes) encourages the combination of DNMTi-based therapies with immunotherapeutic agents. Therefore, further transcriptome analyses of transposable elements, when reactivated, may trigger an interferon-mediated immune reaction, are currently ongoing.
Lübbert:TEVA: Other: Study drug; Celgene: Other: Travel Support; Cheplapharm: Other: Study drug; Janssen: Honoraria, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal